Map the environment
The function doVoro() seen in the previous tutorial is automatically mapping the environment - unless told otherwise. While the codelines shown in this tutorial could be therefore seen as useless, this tutorial develops the concept of local environment.
Once the tessellation has been processed on the system and the instance of the Tessellation class generated, the local environment of the molecules can still be mapped, as seen in this tutorial.
Local environment
Inside a membrane, a lipid A is always surrounded by N lipids Bi. Each of these Bi belongs to a specific phase Pi, which affects its properties and configuration. The distribution of the phases Pi describes the local environment of the lipid A.
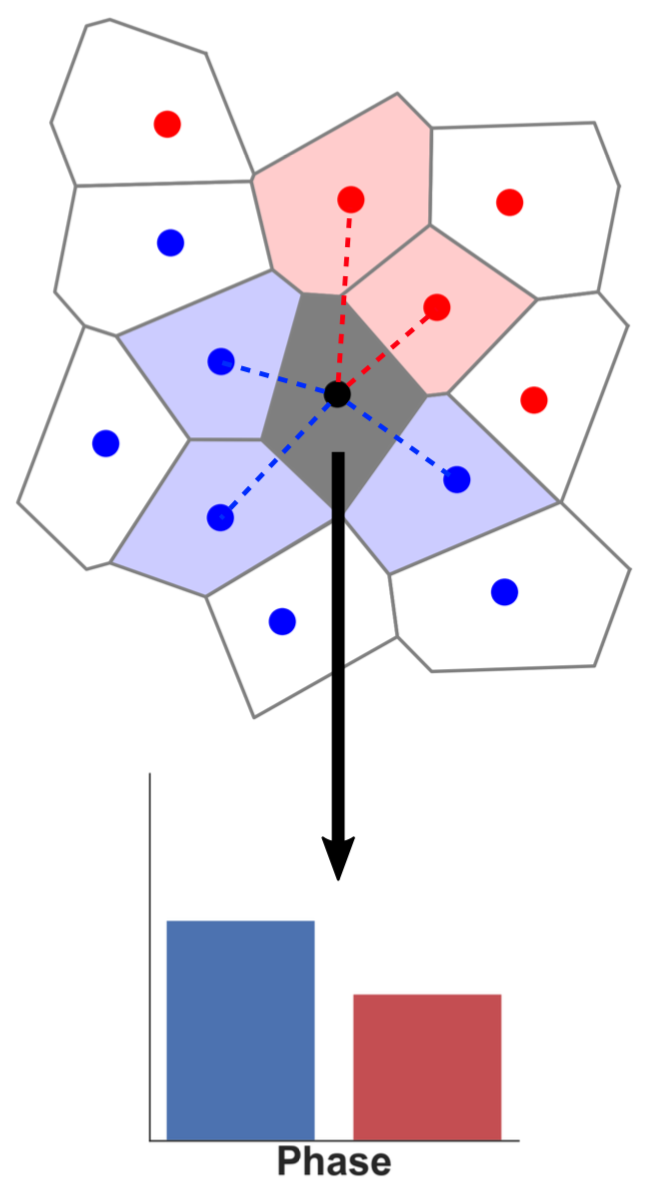
Depending on the distribution of Pi, the location of the lipid on the membrane can be determined:
-
If (almost) all Pi are equal to the same phase Pa, then the lipid is located inside the domain of the Pa
-
If the Pi are distributed (almost) equally between a phase Pa and another phase Pb, then the lipid is located at the frontier between the domains Pa and Pb.
If the total number of lipids in the membrane is large enough, and if the number of lipids in each phase is similar, the difference between the number of lipids found inside a domain and the number of lipids at a frontier can be an accurate description of the state of the phase separation in the membrane.
The mapping of the local environment can be done inside the same leaflet of the lipid A only, or by observing both leaflet at the same time and assuming that the phase on one leaflet is affecting the phase on the other leaflet. The choice between the two observation modes is done when selecting the _3d type of membrane (both leaflet) or not in the doVoro() function.
Run the mapping
The local environment analysis is simply run with the function readNeighbors(), which only takes the instance of the Tessellation class to process as argument.
import mllpa
mllpa.readNeighbors(tessellation)
In this example, the variable tessellation is an instance of the Tessellation class generated via the function doVoro(). Check the related tutorial for more details.
MLLPA will automatically save the results directly inside the instance of the Tessellation class, to be used or saved later. However, it outputs in the same time the total phase composition of each lipid. These values can be stored in variable, such as
neighbor_phases, phases_list = mllpa.readNeighbors(tessellation)
neighbor_phases is a NumPy array of dimensions (# frames, # molecules, # phases). The last axis
reads the number of neighbors found for each of the phases found in the system, e.g. [3, 0, 1] in a system with three phases.
The respective name of the phases corresponding to each index of the list are returned in the variable phases_list
What is next?
-
Now that the local environment has been mapped, the last step is to save all the results in a file.
-
You can also check how to analyse the local environment in a membrane in terms of molecule types and not of phases.
Check the API
The following elements have been used in this tutorial: