Voronoi tessellations
Besides the phase prediction, the other key feature of ML-LPA is its capacity to analyse the local environment of membrane molecules. This is done by performing Voronoi tessellations in order to list the direct neihbors of each molecule.
Defining the geometry
Ghost lipids
Doing a 2-Dimension Voronoi tessellations of a lipid bilayer is straightforward. The two leaflets are studied separately, and for each leaflet all the centers of mass are inside the same Z plane, plus or minus non-significant fluctuations. In this approach, and in this approach only, ML-LPA can proceed straight to the tessellation. However, 3-Dimension tessellations but also alternative membrane geometries, such as lipid vesicles, cannot be processed with the same ease.
In any other type of 3-D Voronoi tessellation, the space is filled with approximatively the same molecular distribution along each axis. When considering a lipid bilayer, the distributions in X and Y, i.e. in the plane of the bilayer filled with lipid molecules, are different from the distribution in Z, filled mostly with solvent and/or solute. If one want to map the lipid neighbors or each lipid, done by discarding all non-lipid molecules from the tessellations, the periodic boundary conditions (PBC) in Z will lead to lipid finding neighbors in the other leaflet by looking through the PBC and not through the membrane. This situation is even worse in the case of a vesicle were lipids are everywhere but always separated by blocks of solvent (invisible during the tessellation). As a result, the neighbor mapping will be significantly biased or even completely wrong.
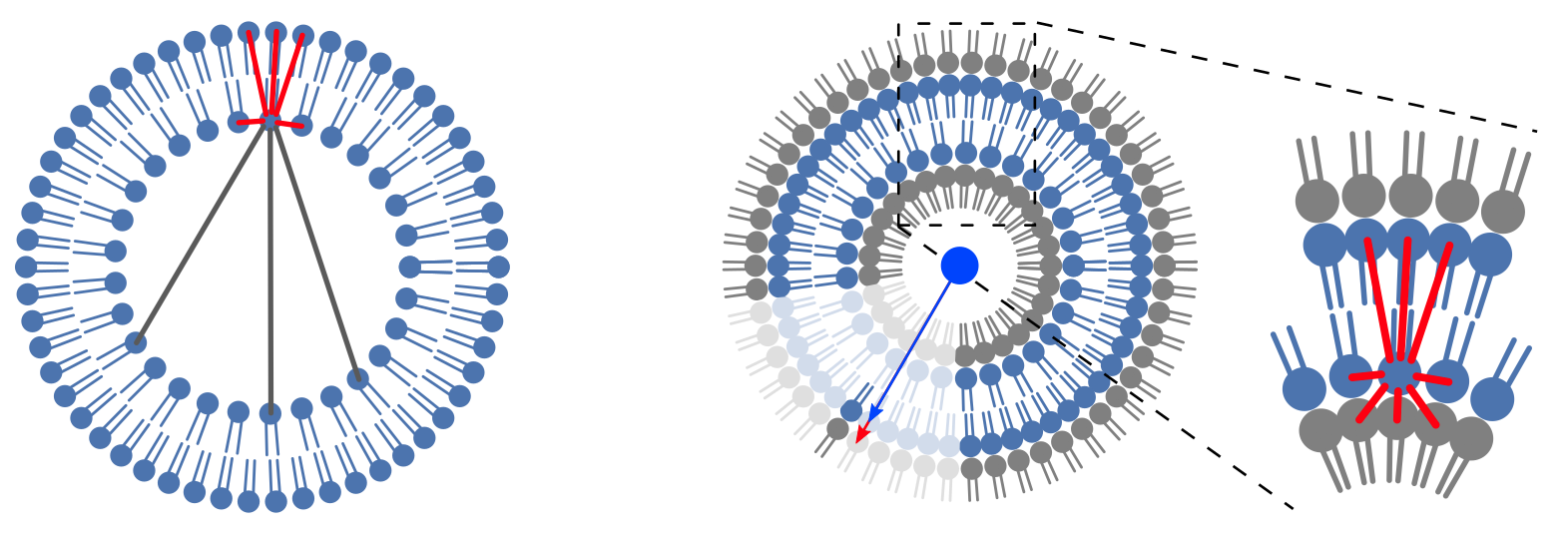
To prevent this, ML-LPA will generate and assign to each lipid a ghost. A ghost is a fake lipid that is created by mirroring the real lipid position at the solvent-bilayer interface. Because of their proximity to the lipid they mirror at the interface, the ghost will always be picked when the tessellation will be searching for the neighbors before the lipid could search through the PBC. Since the ID of the ghost lipids in the tessellation are known by ML-LPA, they are finally removed from the lists of neighbors, living the real lipids with a list on only lipids they detected close to them without going through the solvent.
The membrane geometry
To create the ghosts, ML-LPA will first locate the center of mass of all the lipids of the membrane. Then, it will compute all the vectors between the centers of mass of the individual lipids and the center of mass of the membrane, as well as the distances between the center of mass of each lipid and the position of its furthest atom from the center of mass of the membrane. The ghost is then created by using the vectors and adding to their norm twice the distances. While this process is quite general, it is extremely dependent of the membrane geometry. For instance, the center of mass of a vesicle is easy to define, but a punctual center of mass for a bilayer is meaningless because of the PBC. Instead, for a bilayer, the whole XY plane going through the average Z position of the lipid is used as a center of mass, making all the vectors parallel to the normal to the membrane.
In order to run the tessellation, the user has to specify the geometry. The current version of ML-LPA can work with the following inputs:
-
Lipid bilayers, either with leaflet separation (bilayer) or not (bilayer_3d)
-
Lipid vesicles, either with leaflet separation (vesicle) or not (vesicle_3d)
-
Whole solution without ghost generation (solution)
We are currently working on a new version of ML-LPA to automatically detect the geometry of the membrane. This page will be updated once the detection has been implemented.
Run the analysis
The ghost generation and the Voronoi tessellation are both performed by the same function in ML-LPA, doVoro(). Similarly to the function generateModel(), doVoro() takes as an input a list of instances of the System class. However, if only one instance is given, doVoro() will automatically read it as a list of Systems. As explained above, we also need to specify the geometry of the membrane.
import mllpa
tessellation = mllpa.doVoro(unknown_system, geometry='bilayer')
In this example, we only wanted to analyse one instance of the System class, unknown_system. All the relevant information on the systems and their tessellations have been stored in the variable tessellation as an instance of the Tessellation class. More information on the Tessellation class are given in the API.
Since doVoro() needs to calculate the center of mass of the whole membrane, it is essential to include all the systems containing all the molecules making the membrane.
In some cases, it can be important to exclude some molecules from the ghost generation. For instance, molecules completely inserted in the hydrophobic tails of the lipids (e.g. cholesterol) do not need ghosts, and having ghosts might create issues when the Voronoi cells of the individual lipids will be generated. To prevent this, we can exclude some systems from the ghost generation using the exclude_ghost= keyword-argument. It takes as an input a list of indices corresponding to the systems to exclude in their respective input list.
ternary_tessellation = mllpa.doVoro([unknown_dppc, unknown_dopc, unknown_cholesterol], geometry='bilayer_3d', exclude_ghost=[2])
In this example, we exclude the 3rd system ([2]), corresponding to unknown_cholesterol from the ghost generation.
doVoro() has been programed to automatically analyse the local environment and map it. This can be disabled by using the keyword argument read_neighbors= and set it to False.
tessellation = mllpa.doVoro(unknown_system, geometry='bilayer', read_neighbors=False)
Advanced notions
Spurious neighbors
The main use of the Voronoi tessellation in ML-LPA is to find the closest neighbors of each lipid. However, due to the complexity of a 3-d tessellation, geometrical aberrations can occur. As a result, two molecules that are not closest neighbors will be identified as such because the tessellation was able to find even a micro-surface that could connect both.
To remove such spurious neighbors from the list of neighbors, ML-LPA apply a threshold at a given ratio of the total surface of the Voronoi cell. Each facet connecting two molecules being below this ratio will be removed from the list. By default, the threshold value is set to 1% of the total surface of the cell. It can be changed with the keyword-argument threshold=.
Area and volume per lipid
Performing the Voronoi tessellation automatically computes the volume of each of the Voronoi cell. For a lipid bilayer tiled in 2-D, this corresponds to the area per lipid. For a bilayer tiled in 3-D, this is the volume per lipid.
This values can be accessed in the instance of the Tessellation class generated by doVoro(), by using the .volumes attribute.
lipid_area = tessellation.volumes
Both lipid areas and lipid volumes are called from the .volumes attribute. The nature of the value extracted depends on the geometry selected in the doVoro() keyword-argument.
What is next?
-
Now that the system has been tessellated, you can use the tiling of the space to map the local environment.
-
You can check how to analyse the local environment in a membrane in terms of molecule types and not of phases.
-
You can also save the results in a file.
Check the API
The following elements have been used in this tutorial: